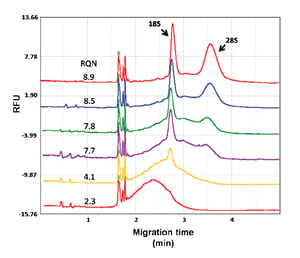
Library preparation is the most critical step in the NGS (next generation sequencing) workflow. The preparation of DNA fragments of specific size ranges is the basis of high-quality analyses. Therefore, monitoring the sample size and quality of the NGS library is of paramount.
The following publication (read more >>>) demonstrates how to use the fully automated Qsep100 Biofragment Analyzer (read more >>>) is used to detect smallest NGS library samples (up to a limit of 50 pg). The Qsep100 system was used to monitor each step of the NGS library preparation workflow, from upstream genomic DNA, to total RNA to the final step of the DNA library.
The Qsep100 analyzer showed through e.g.,: easy use + innovative microcapillary electrophoresis technology with ready-to-use and replaceable gel cartridges + high resolution and high throughput + reduced cost of the instrument and also the costs per sample analysis.
Next-generation sequencing (NGS) is widely used in genomics, epigenetics and transcriptomics, etc.,1 and also in such clinical diagnostics applications as cancer screening and drug development.2 There are multiple commercial NGS platforms available, each incorporating unique operating principles (e.g., Roche 454FLX [454 Life Sciences, Branford, Conn.], Illumina Genome Sequencer3 [Illumina Inc., San Diego, Calif.] and Applied Biosystems/ABI SOLiD Sequencer [Foster City,Calif.]). No matter the platform, library preparation is the most critical step in the NGS workflow.

References
-
Metzker, M.H. Sequencing technologies—the next generation. Nat. Rev. Genetics 2010, 11, 31–46.
-
Desai, A.N. and Jere, A. Next-generation sequencing: ready for the clinics? Clin. Genetics 2012, 81, 503–10.
Quail, M.A.; Kozarewa, I. et al. A large genome center’s improvements to the Illumina sequencing system.
Nat. Methods 2008, 5, 1005–10.
Qsep Bio-Fragment Analyzer

